By understanding the details of how bacterial genes are controlled, we can develop new approaches that interfere with the processes and kill deadly bacteria.
A dedicated protein called RNA polymerase (RNAP) moves along DNA copying its information into a strand of RNA. This RNA then directs the production of the proteins that build and control most of the processes in all living cells. However, sometimes as the RNAP moves along the DNA, it gets stuck and blocks other processes, potentially leading to cell death.
The researchers found that another protein called HelD removes and recycles stuck RNAP from the DNA. Now, by using cryo-transmission electron microscopy at the Microscopy Australia Linked Lab at the University of Wollongong, the researchers have worked out how HelD does it in the bacterium, Bacillus subtilis, which is related to dangerous human pathogens Bacillus anthracis (anthrax) and Clostridium difficile. The Czech collaborators discovered a variant of the same mechanism in mycobacteria, which cause tuberculosis and other devastating diseases.
The cryo-TEM revealed that HelD has powerful ‘arms’ that reach deep into the RNAP to prise it open and clear away all the residual material that had been blocking it up. HelD, then resets the RNAP structure so it can start working again.
This previously unknown mechanism for RNAP rescue and recycling leads the way to development of new drugs to fight bacteria.
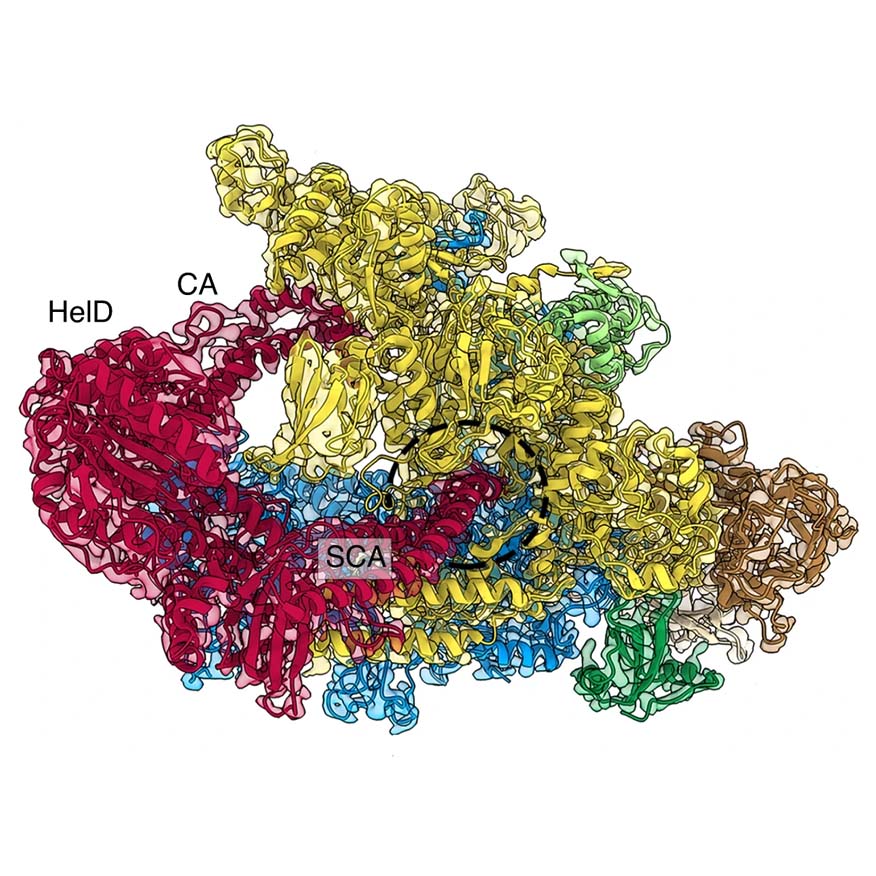
Reconstruction from cryo-TEM data showing the structure of HelD (red) interacting with RNAP.
T. P. Newing, et al., Nature Communications 2020 DOI: 10.1038/s41467-020-20157-5
Array
January 21, 2022